Discovery health information model
Information modelling is the set of processes by which representations of data relationships are created maintained and queried.
The Discovery models are designed both for human visualisation and for computers to use.
Systems that use the models can use any or all of three approaches:
- Direct use of the model data content as a database (or set of files that can populate a database via script)
- Use via a set of APIs (both local and remote) designed to provide access to the data within the model, or to trigger outputs of the model for 1)
- Use of the information model technologies themselves via the use of the published open source code
Information model functions
The information Information models have 4 core functional requirements internal to the model: Description of the model , validation of model content, population of the model, and query of the model. In support of query there is also the need to support inference which generates new insights that were not necessarily authored.
In addition the information model must support the same 4 core functional requirements on actual health data that is modelled.
- Description of the model. There is little point in having a model unless it can be described and understood. Knowing what is in a model is a pre-requisite to using it. For example, there is no point in trying to find out if a patient record indicates whether or not they have diabetes if the model doesn't include the ability to record it. In order to understand a model, two techniques are required: diagrammatic representation and human readable text representation. A model must support both.
- Data Validation is essential for consistent business operations. Data models, user input forms, and data set specifications are designed to enable data collections to be validated. Maintaining a standard for data collection is essential. For example, if you have a patient record in front of you, you will likely need to know their approximate age. To work this out date of birth must be recorded. Validating that the date of birth can be and has been recorded is important. However, if more than one date of birth was recorded for the same patient, it would be less valuable. Thus a modelling language must include the ability to constrain data models to suit particular business needs.
- Population of the model. It is impractical to build model content from scratch and likewise virtually impossible to populate instances with existing data without some manipulation. An information model must contain the ability to model mappings between currently held data and model conformant data.
- Enquiry (or query) is necessary to generate information from data. There is little point in recording data unless it can be interrogated and the results of the interrogation acted upon. Thus a modelling language must include the ability to query the data as defined or described, including the use of inference rules to find data that was recorded in one context for use in another.
- Inference is pivotal to decision making. For example, if you are about to prescribe a drug containing methicillin to a patient, and the patient has previously stated that they are allergic to penicillin, it is reasonable to infer that if they take the drug, an allergic reaction might ensue, and thus another drug is prescribed. Thus a modelling language must include the ability to infer things and classify things for safe decisions to be made
Model structure
A model must be built from some structure, using some tools or processes to build it. This section describes the nature of the structure that makes up the information model. The tools used to build the model includes the use of an information modelling language which is described separately.
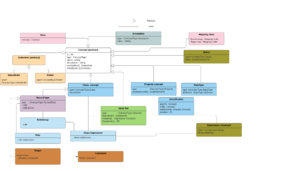
A model must have a model i.e. a meta model that models the model i.e. the types of things a model is made up of.
The Discovery model can be described as an "Object Role Model (ORM) that includes an Ontology as one of the roles". It can also be described and implemented as a small number of main classes with each main class covering a role type.
Both perspectives
The roles themselves can be categorised into types. As one of the types of roles includes ontological axioms, this means that the model can operate both with the open world assumption (as required by the semantic web) and a closed world assumption (as required by the business of healthcare).
The main types are illustrated in the right hand image.
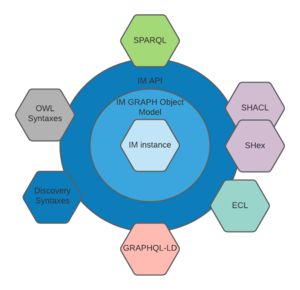
Interaction between the model and the external world is undertaken via the Discovery information model language, (or alternatively a set of W3C recommended languages) . These are described separately but consists of a language built from RDF triples applying the W3C language grammars and vocabularies of OWL2, SHACL, SPARQL, with support for GRAPHQL
The following sections briefly describe the various model classes illustrated above. The IM language specification provides more insight into the details of the classes.
Concept
All things that can be referenced via an identifier can be thought of as a concept. Even the classes and structures of the information model themselves are concepts.
A concept is defined as an ‘abstract idea’ or ‘general understanding of something’ and this meaning is preserved in the modelling language. It is one of the few abstract classes in the information model. This means that there is no actual object of 'type concept' unless it is also a type of some subtype of a concept.
Types of concepts include : Class, Property, Shape, Value Set, Data type, Query, Collection, term, and annotation. Each of these specialise in their function and properties and inherit the core properties of a concept and specialise by extension.
Use of aliases. Aliases enable properties and classes to be used in their alias form. See language specification for how context is used to provide aliases to enable key terms to be used in business processes without the inconvenience of using IRIs. Thus these sections use aliases for convention, the aliases themselves defined as aliases to concepts.
A concept also comes with a fixed set of annotation properties that can be relied on to be present or have null values
| Property | Type | Description |
|---|---|---|
| iri | IRI | an international identifier, the format as described within the language specification |
| status | enum | A status concept representing the status of the concept in terms if its activity status e.g. Active or inactive |
| name | String | This is the full name of the concept (or preferred term in Snomed-CT.) In OWL2 this is a label annotation |
| description | String | A plain language meaning of the concept, and how it may be used |
| version | integer | The version in which this concept was first created |
| code | String | If the concept has a code, the code assigned to this concept by the original creator, e,g, a Snomed-CT, READ2, ICD10, OPCS or local code or auto generated code |
| scheme | IRI | f the concept has a code, the code scheme assigned to this code, the scheme itself being an IRI |
| termKey | String[] | A number of keys used to link to the concept. Should not be confused with a term concept which is an alternative term linked to a concept |
| annotation | Annotation[] | Concepts may have additional informative simple string properties used for a variety of business purposes |
| alias | String[] | Aliases for this concept i.e. reserved terms within the context of a particular application that is implementing the information model and wishes to use aliases rather than the IRIs |
Ontological Class
An ontological class is an extension of the concept class and is used as the main means for defining semantic concepts for use in healthcare records.
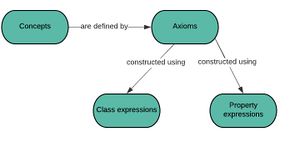
The difference between an ontological class (often referred to as an owl class) and a simple concept is that it can be semantically defined by the use of class axioms. Class axioms such as subclass or equivalent classes are used for reasoning and enable the information model to be queries using subsumption query.
Information model storage architecture
An information model is an abstract representation of data, but an information model must have content and that content must be stored.
Data cannot be stored conceptually, only physically, and thus there must be a relationship between the abstract model and a physical store.
In the information model services, the abstract model is instantiated as a set of objects of classes, the data element of those classes holding the subject, predicate and object structures. In reality those objects together with translation and data access methods are instantiated in some form of language. e.g. Java.
The physical store is currently held in a triple like relational database accessed by a relational database engine but could be easily stored as a native graph.
The model can then be used as the source and target of the exchange of data, the latter using a language interoperating via a set of APIs
This can be visualised as in the diagram on the right. It can be seen that the inner physical store, is accessed by an object model layer, which is itself accessed by APIs using modelling language grammar and syntax. The diagram shows the main grammars supported by the Discovery information model, including the Discovery information modelling language grammar itself.
Support for the main languages means that a Discovery information model instance has 2 levels of separation of concerns from the languages used to exchange data, and the underlying model store. There is thus no reason to buy into Discovery language to use the information model.
Likewise, an implementation of objects that hold data in a form that is compatible with a particular data model and ontology module, can be accessed using the same language.
This makes the language just as useful for exchanging query definitions, value sets as well as useful for actual query of health record stores via interpreters.
The remainder of this article describes the language itself, starting with some high level sections on the components, and eventually providing a specification of the language and links to technical implementations, all of which are open source.
Model structures
The information model is made up of several types of structures. There are various ways in which the structural types can be classified but broadly speaking can be divided as follows.
Semantic Ontology
All concepts, wherever used, are defined in the semantic according to their meaning via constructs known as axioms. The definitions themselves are created and maintained by the ontology authors e.g. Snomed International or NHS Digital. A modular approach is used whereby concepts are shared across modules but modules may create new axioms to further refine the definitions as long as they are consistent with the authored definitions. Examples of a semantic ontology modules are Snomed-CT ontology, PROV (provenance) , and the Discovery semantic ontology (which in essence is a Snomed extension).
The ontology also contains concepts from other non ontological sources, including standard classifications. Where different concepts exist that mean the same thing they are either resolved to a single concept, or asserted as equivalent. Where external concepts that are not defined are entailed by a core concept they may be asserted as subclasses. If it unclear then a mapping relationship is established.
As mentioned the ontology consists of a modular ontology which is a super ontology made up from ontology modules, which may often be also separate ontologies in their own right
Classification
The models include modular classifications of concepts. The classification modules are either generated from the ontology via classifiers (which are functions of reasoners) or have been incorporated as handcrafted classifications . Examples of ontology generated classifications are Snomed-CT "ISA" hierarchy and the Discovery health classification. Examples of handcrafted classification modules are ICD10, Read. The main thing to note about the difference between the two is that concepts in classifications that are generated from an ontology subsume their descendants as proper subtypes whereas handcrafted classifications may include subcategories that are inconsistent.
To illustrate the difference between an ontology and a classification, Let us say that we state in an ontology that "ALL THINGS THAT BARK ARE DOGS".
Let us then say we go to the beach at Ravenscar on the North East coast of England and hear a bark. We see the animal at a distance. We ask the computer what it is. The computer, using the generated classification would classify that animal automatically as a DOG ( because it barks) .
However, as we get closer we see that it is something else. The ontology is clearly incorrect. Consequently we amend the ontology to state that there is such a thing as an "AN ANIMAL THAT BARKS" and that "A DOG IS A SUBCLASS OF AN ANIMAL THAT BARKS". We also state that such a thing exists as an "ANIMAL" and that "AN ANIMAL THAT BARKS IS A SUBCLASS OF AN ANIMAL". Now, when asking the computer what the animal is, the computer knows only that it is an animal that barks but does not know what it is.
We then amend the ontology to state that such a thing exists called a SEAL and that a "SEAL BARKS". We also author the ontology to say that "AN ANIMAL THAT BARKS IS EQUIVALENT TO A THING THAT BARKS" i.e. by definition if it barks it is an animal that barks. Now the computer automatically classifies the seal to state that "A SEAL "IS A" ANIMAL THAT BARKS" (because it is a thing that barks and must therefore be an animal that barks). It will then be found when searching for types of animals that bark, things that bark, and things that are animals i.e. the seal MUST be an animal because all things that barks are animals that bark and an animal that barks is a subclass of an animal. The human has not needed to find the category, the reasoner does it for you, it has automatically created the classification from the properties of the thing on the beach.
Data models and value sets
Business domain data models are modules that define relationships in the context of a particular business or set of businesses and include the health data models.
A model is only relevant for a particular set of business purposes and here is no single model that can accommodate all business purposes, although common information models can accommodate quite broad purposes. A reasonably well understood set of business purposes is referred to in these topics as a "business domain" or "domain of interest" and a particular information model is designed to cover a business domain.
Examples of data model modules are the core Discovery health data model, and the PRSB core record model. Related message models such as FHIR profiles or openEHR archetypes are examples of business domain specific data models. Specialist data models may exist for particular business purposes such as cancer data set definitions or be more general, such as the Discovery common data model. The thing to note about the Discovery data models is that all concepts are defined in the semantic ontology.
The Discovery common data model (a broad model for each domain) will generally include data relationships needed by many domains, arranged in a way that inconsistency or unreliability is avoided.
Data models, also define the expected values of properties. Sometimes these values are class statements (e.g. has colour -> Colour meaning that the colour of something is a colour) and more often they are sets of concepts brought together for business purposes - value sets.
Query Library
A variation on a conventional ontology is that concept properties can also be defined according to functional definitions, as expressed in query language. The model contains a library of query definitions that like Data models, are usually business specific.
For example one would expect a record of a person's religion to be the concept of "Person's religion". In a data model this might be defined as " Person-> has religion -> Religion i.e. the value of the religion property of a person is a religion e.g. Hindu.
However, the same person may have many religions recorded about them during their lifetime. Thus the definition of a person's religion is more likely to be "the latest religion for this person" and that is a functional property.
A query is essentially a definition of a concept using both standard and functional properties, together with the related value sets with the addition of instructions as to what properties to return.
The model building bricks
All the Discovery models, whether semantic models or business related data models are built using the same machine readable language. More specifically, the language grammars used are based on the following set of open standard based languages, interpreted in a way that matches the businesses:
The language(s) are further described in the article on Health Information modelling language.
Ontologies and modules
The Discovery common information model can be thought of as an ontology of ontologies. More precisely though it should be considered as an ontology consisting of a set of ontology modules with each module defined according to business needs. The principle of concept sharing, whereby one concept is identified once across the entire set of domains, suggests that there is a single ontology. However a data model that is specified for a particular business purpose may have different class structures from another business purpose even though they share the same semantic definitions.
For example, take the idea of recoding information about a blood pressure. This is an example of a component in a data model. In General practice, it would be common practice to record a systolic and diastolic blood pressure and thus the component would consist of 3 classes. However, in a specialist research study involving different interpretations of blood pressures, including perhaps the size o nature of the cuff, or the exact position of the patient, this component may be more complex.
This is addressed by modularisation where the axioms that define the classes belong to a particular model, even though the property domains and their ranges are shared across the ontology. This is analogous to the idea of templates derived from subsets of archetypes. The difference is that there is no "super-archetype" requiring international agreement on the items in the archetype, but instead there is a demand that the same identifier of the diastolic blood pressure record class is used throughout, even though the class definition is business specific.
Disambiguation of terms
Throughout the healthcare domain, the same terms are often used to mean different things when used in different contexts. This can create some fundamental problems in design, which the information modelling attempts to overcome.
Take a "blood pressure" as an example, this could be used mean :
a) The actual blood pressure observation itself (a blood pressure was observed)
b) The record of a blood pressure (Mrs Smith's blood pressure on 1st February),
c) The blood pressure procedure (A blood pressure procedure was undertaken).
This ambiguity is addressed by using different concept identifiers for the different contexts. Editorial policy would normally disambiguate the term e.g. blood pressure (observation), blood pressure measurement (procedure), or blood pressure (record).
The net result of disambiguation is the generation of many more identifiers for record based classes in the model. In the blood pressure example several identifiers are likely to be used to model the blood pressure related in the following way:
In the GP Domain Module a blood pressure data model might be:
Blood pressure (record) -> is a subclass of -> observation (record)
& -> is a record of -> a blood pressure (observation)
& -> has subcomponent- > [of systolic blood pressure (record),
and diastolic blood pressure (record)]
This creates a type hierarchy in the data model similar to the type hierarchy in the semantic model. From an implementation perspective it is likely that the "Observation" would be mapped to a table directly, whereas the blood pressure record would be assigned to a "record of" or "type" column, the content of which could be any object that is a member of the observation type value set, and 2 additional observations as components would be created. HL7 FHIR uses this model via the use of the generic term "code" and "component".
Unlike other frameworks such as FHIR or OpenEHR there is no health specific "Reference" model as such beyond the OWL standard need to differentiate classes from properties from data types. This is because a selected structure from the top down is determined by interpretation, use case and the communicating community.
Models viewed in packages
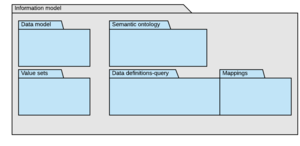
Another way of categorising the information models is by the use of the idea of packages.
Discovery information models can be said to reside within one of 5 packages, each package directed at particular sets of use cases. Two of the packages fall into the sematic ontology domain type, and 3 fall into the data modelling domain type.
Crucially, all the packages are integrated by a common language and share the same concepts, each of which are defined within a semantic ontology.
- The semantic ontology is the set of concepts used in all parts of the information model, from clinical concepts through to data structure concepts
- The data model is a set of entities, attributes and value sets, all of which are defined precisely in the ontology, but he data model, being created for a specific business of healthcare is separate to the ontology.
- Value sets , or concept sets, are business purposes specific collections of concepts from the ontology used in the data model or in query and contain concepts as defined in the ontology, using the ontology language, including advanced concept classes.
- Data set definitions apply rules and filters to a data model in order to specify the nature of the entries and their content required in a purpose specific data set
- Model maps specify how data is transformed from a data model to a particular database or messaging format.
- Data base schemas are reference schemas (RDB and maps) showing an implementation of a data model and data sets. Strictly speaking these are not part of the information model but are included as “proof of solution” of the model.
- Query definitions are a library of re-usable queries.
Semantic ontology
A semantic ontology defines the meaning of the concepts that make up the content of health records. The meaning is defined in a way that a computer can use to classify, reason and analyse.
As a semantic ontology is based on Description Logic, the semantic ontology applies the same interpretation to its concepts for classification as does OWL However when used for purposes such as authoring and value set generation, closed world assumption is used of the kind described in the Snomed expression constraint language.
The world leading Snomed-CT ontology forms the major part of the semantic ontology for the definition of health characteristics, supplemented by maps to the legacy concepts such as ICD10 o Read codes. Consequently the semantic ontology can be represented in several syntaxes including the mainn OWL syntaxes, Manchester Syntax, Snomed compositional grammar and Expression constraint language
An ontology is made of of a number of axioms which relate concepts to other concepts in a fractal like manner. T
The ontology may also be defined using the Discovery semantic ontology language, which is itself a syntactical simplification on the standard OWL2 language. The Discovery language exists in order to accommodate additional constructs not covered in OWL, namely data set definitions, value set definitions, and transactional messaging.
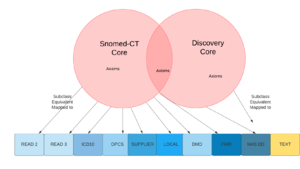
Relationship between core and legacy
The semantic ontology can be modularised into Core and Legacy concepts according to the namespaces of the concept identifiers. That is not to say that the legacy concepts are not used. Quite the reverse. Given that 99%+ of all healthcare data is still recorded using legacy concepts the semantic ontology must incorporate these. In addition a vast number of system specific or provider specific codes are in use.
Both core and legacy are defined by OWL2 DL axioms. However, the core concepts are likely to be defined in a way that sufficiently identifies the concept within the domain whereas legacy concepts are more likely to be defined only to the extent necessary for query subsumption. Nevertheless legacy concepts may also be sufficiently defined. In many cases those definitions come in the form of expressions containing core concepts.
For example, An adverse reaction to Atenolol is a GP legacy concept. This can be sufficiently defined in the following Axiom
Adverse reaction to Atenolol - is equivalent to - (Adverse drug reaction & causative agent = Atenolol)
The Discovery common model creates relationships between core and legacy using a mapping relationship, the commonest being
- Equivalent. Where the legacy code or term is deemed to be equivalent in meaning and definition to the core concept
- Subclass. Where the legacy code or term is deemed to be subclass of the core concept
- Mapped to. Where the legacy code would be expected to be a member of the set defined by the core concept, but may not be sufficiently defined to be confident of equivalence or subclass.
From a mapping perspective the maps operate from Core -> Legacy and not the other way round. For example, if one were searching for Diabetes using a core concept, and a patient had a diagnosis of the ICD10 code "Diabetes without mention of complication" then one would expect that patient to be found (depending on the enquirers preference). However, if querying on "Diabetes without mention of complication" then no core concept would be found as the relationship does not go forward. The exception to this rule is the "equivalent" axiom which is bidirectional.
If the relationship between a core and a legacy is "equivalent" or "subclass", this does not mean that the child codes of the legacy codes would normally be included, as the child codes are often not subclasses from a semantic perspective. This is important to recognise when authoring queries using core concepts, operating on data that uses legacy codes.
Value sets or concept sets or reference sets
Main article : Value_sets
A value set definition, and it's run time counterpart- value set transitive closure , is a set of class expressions collected together for a particular business purpose.
There are a range of purposes for a value set. Examples range from defining a data set according to a set of recorded concepts, indicating the expected range of a property in a health record, or testing the presence of a feature in a patient record.
Data models
Main article : Data Model
A data model is module of an ontology that defines classes required for particular business purposes.
Business purposes vary from the need to store particular items of data through the need to display items in a certain way. This is the model that defines the ever evolving structure of health records held within multi-domain health records, varying from common high level classes through to specialised classes. An example of the former is an 'observation', and an example of the latter is a 'Blood pressure' or an 'histological/immunological report on a breast carcinoma'.
N.B. in IS013606 these are called archetypes and their derivative templates. In FHIR they are referred to as resources and profiles.
Data definitions - query
Data set definitions or queries are a key component of the information model.
A data set definition is a specification of a subset of data derived from one or more data models
A data set definition, once established, can also be used as a source data model and thus data sets can be chained by placing a data set into the role of a data model.
A data set uses query like constructs to define its structures. Data set entities and data set attributes may be derived from a combination of ontology and data model query. To that extent, a data set definition can be said to use a query language.
The Discovery data definition language is not designed to operate as an actual query language, as it does not extend to include all the sophistication needed by a run time query language. For example, there are no optimisation techniques employed or references to the use of indexes. However, the language is sufficiently rich to be able to easily generate SQL or Cypher from the specification when used with a data model map to the implementation schema.
Data Maps
Data maps hold the maps for a variety of purposes, mainly being:
- Maps between the data model and an implementation schema to enable auto generation of query syntax such as SQL or CYPHER
- Maps between legacy data models and or their values to the common information models