Inbound Data Transform Specifications: Difference between revisions
No edit summary |
|||
| Line 30: | Line 30: | ||
{{Note| the image represents the logical flow of data into the DDS, rather than the distinct software components themselves.}}<br /> | {{Note| the image represents the logical flow of data into the DDS, rather than the distinct software components themselves.}}<br /> | ||
== Publishing process == | <br />== Publishing process == | ||
Revision as of 08:29, 22 April 2021
Introduction
This article forms the basis of the technical implementation of any new inbound transform of data into the Discovery Data Service (DDS), specifically for non-transactional flat file formats.
The aim is to ask potential publishers to consider and answer the following questions:
- How will the source data be sent to DDS? Will it be an SFTP push or pull? Will the files be encrypted? See Secure publication to the DDS for currently supported options.
- How often will the source data be sent to DDS? Will it be sent in a daily extract? See Latency of extract data feeds to see how often and when data is currently received, and how that relates to the data available in the DDS.
- How should the source data be mapped to the FHIR-based DDS data format? What file contains patient demographics? What column contains first name? What column contains middle names? See Publishers and mapping to the common model for more information.
- How should any value sets should be mapped to equivalent DDS value sets? What values are used for different genders and what FHIR gender does each map to?
- What clinical coding systems are used? Are clinical observations recorded using SNOMED CT, CTV3, Read2, ICD-10, OPCS-4 or some other nationally or locally defined system?
- How are source data records uniquely identified within the files? What is the primary identifier/key in each file and how do files reference each other?
- How are inserts, updates and deletes represented in the source data? Is there a 'deleted' column?
- Is there any special knowledge required to accurately process the data?
- Will the publishing of data be done in a phased approach? Will the demographics feed be turned on first, with more complex clinical structures at a later date?
- What files require bulk dumps for the first extract and which will start from a point in time? Will a full dump of organisational data be possible? Will a full dump of master patient list be possible?
- Is there any overlap with an existing transform supported by DDS? Does the feed include any national standard file that DDS already supports?
The understanding of the answers provided can then be used by the DDS technical team to implement the technical transformation for the source data.
Completion and sign off of the specifications is a collaborative process, involving the data publisher (or their representatives) and the DDS technical team; the data publisher has a greater understanding of their own data, and the DDS technical team have experience of multiple and differing transformations.
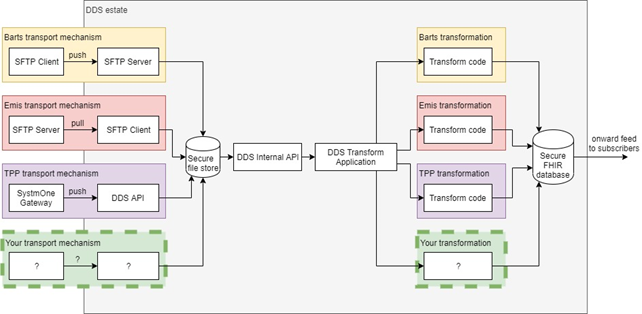
DDS Inbound Overview
This diagram provides an overview of the way the DDS processes a sample of existing inbound data feeds. Each data publisher has their own transport mechanism for getting data into the DDS estate (SFTP push, SFTP pull, bespoke software) which is then sent through a common pipeline that performs a number of checks (most importantly validating that DDS has a data processing agreement to accept the data). Once the checks are passed, the data is handed over to the DDS transformation application, that then invokes the relevant transform code for the published data format. The result of the transformation code is standardised FHIR that is saved to the DDS database; this may be further transformed for any subscribers to that publisher’s data.
Unless there are exceptional circumstances that preclude it, all DDS software is written in Java, running on Ubuntu.
| the image represents the logical flow of data into the DDS, rather than the distinct software components themselves. |
== Publishing process ==